
Modern transformation of ancient healing arts: traditional Chinese medicine in cancer immunotherapy
Introduction
The immune system, under normal circumstances, maintains a delicate equilibrium between the Yin and Yang of immune tolerance and autoimmunity (1). Insults at the cellular, genetic, and epigenetic levels tend to induce the tissue’s physiological responses, including cell proliferation, migration, and inflammation, and facilitate either tissue repair or scarring (2-6). One of the key events leading to tumorigenesis is the disruption of immune homeostasis, in which abnormal cells accumulates mutations and epimutations (or epigenetic alterations) to evade immune surveillance, leading to uncontrolled proliferation and immortalization of abnormal cells (7,8).
The intricate network of immune cells orchestrates a series of mechanisms to protect the host from pathogens and diseases, and these mechanisms are dichotomized into two lines of defense: innate immunity provides immediate but nonspecific responses against threats before the more specific adaptive immune response is initiated (9). Natural killer (NK) cells and CD8+ T-cells (cytotoxic T lymphocytes, CTLs) are the most potent antitumor power to recognize and eliminate cancers. NK cells of innate immunity are responsible for cancer immunosurveillance without prior sensitization (10). Major histocompatibility complex class I (MHC-I) downregulation is a significant mechanism exploited by many types of solid tumors to evade immune detection by the adaptive immune system (11-13). NK cells specifically recognize a reduction in surface MHC-I presentation, determining those cells as abnormal and eventually sending them for destruction (14). In addition to NK cells from innate immunity, CTLs from adaptive immunity also have the capacity to kill tumor cells directly. CTLs scrutinize all nucleated cells in the body via the interaction between T-cell receptors (TCRs) and antigen peptide/MHC-I complexes. CTLs induce apoptosis upon the recognition of peptides presented by MHC-I molecules on target cells (15).
During the past decades, immunotherapy has revolutionized cancer treatments and established the theoretical grounds to develop more effective treatments (16). Despite these promising discoveries, tumor cells are intelligent in developing resistance by manipulating their microenvironment and inducing anti-tumor immune cells anergy, rendering immune checkpoint blockades (ICBs) ineffective in many types of cancer (17). In this review, we briefly discuss CD8+ T-cell biology and tumor immune evasion mechanisms, assess the significance of antigenicity loss in solid tumors and list ongoing efforts in enhancing antigenicity in solid tumors. We further propose several future research directions on the immunoregulatory effects of traditional Chinese medicine (TCM) as potential novel therapeutic strategies.
T cell activation and killing mechanisms
Naïve CD8+ T-cells develop and mature in the thymus. These naïve cells enter the bloodstream upon maturation and circulate through the secondary lymphoid organs (15,18). They can encounter potential antigens presented by the MHC-I on antigen-presenting cells (APCs) such as dendritic cells (DCs), macrophages, and B cells (19). T-cell activation requires at least three signals: an initial antigen-specific signal, a co-stimulatory signal, and cytokines (20,21). All events must occur sequentially to induce adequate T-cell activation and response. First, the TCR of a naïve CD8+ T-cell recognizes an antigen presented on the MHC-I of an APC. A CD8 co-receptor then stabilizes the initial interaction between TCR and MHC-I. After forming a stabilized complex, the expression of B7.1 or B7.2 by APCs is induced, allowing the critical second B7-CD28 co-stimulatory signal to fine-tune the T-cell response. Lastly, IL-2 production by the activated CD8+ T-cells, or more often, with the help of CD4+ effector T-cells, promotes differentiation into CTLs that directly kill foreign or cancer cells (22).
The duration of antigen exposure is critical to maintain effective CTLs. Antigens need to be present and recognized to trigger CD8+ T-cell differentiation. The host must be able to rapidly eliminate such antigens from the system after the maturation of CTL to allow the development of memory CD8+ T-cells. Memory CD8+ T-cells are powerful in fighting off foreign substances upon re-exposure. In cancer and chronic viral infections, when antigens are not removed from the host promptly, neoantigens are sampled and circulated constantly. This prolonged exposure to antigens leads to desensitization and impaired memory CD8+ T-cell development, resulting in CD8+ T-cell exhaustion (23-29). The key features of exhausted CTLs are loss of effector functions, sustained expression of inhibitory receptors, altered transcriptional and epigenetic modifications, and metabolic reprogramming (Figure 1) (23). Thus, to restore CTLs activity and functions, therapeutic strategies must be considered to tackle such complex tasks at hand.
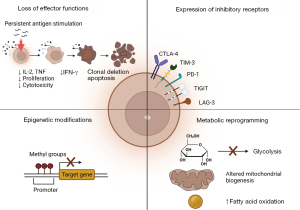
CTLs have been a primary focus in immunotherapy research due to their specificity and high efficacy in targeting tumor cells that carry specific antigens (29-33). Antigen-MHC-I complex recognition by TCR triggers the release of two populations of cytotoxic granules, inducing caspase 3-mediated apoptosis through two distinct mechanisms (34). One granule population of CTLs contains granzymes, perforins, and granulysins. The released perforins create pores on the target cell membrane, thereby delivering granzymes that activate caspase 3 intracellularly, leading to target cell apoptosis. The other granule population of CTLs consists of Fas ligands (FasLs), which activate caspase 3 by engaging the Fas receptors on the targeted cell surface. The FasL/Fas receptor interaction results in caspase 8 activation. Activated caspase 8, in turn, activates caspase 3, inducing apoptosis. TCR binding also modulates cytotoxic molecule replenishment via de novo synthesis, allowing a CTL to kill a series of targets in succession (35). Besides their direct killing capacity, CTLs release several cytokines, including IFN-γ, TNF-α, and TNF-β, which facilitate tumor cell recognition and elimination (36). Most importantly, IFN-γ has gained much attention in immunotherapy development due to its ability to enhance MHC-I expression and antigen loading in tumor cells (37-39).
Tumor immune evasion mechanisms overview
Cancer immunotherapy is designed to augment the immune response against tumor cells. Strategies such as ICBs and adoptive T-cell therapy have revolutionized cancer treatment (16,40,41). While ICBs have shown extensive clinical success in treating melanoma and non-small cell lung carcinomas, their effectiveness in treating many other solid tumors is limited (42-49). Tumor immune evasion through immunoediting and tumor heterogeneity are two mechanisms that are responsible for the observed suboptimal response to ICBs in many patients (50).
Tumor immune evasion poses a significant barrier to treating solid tumors. Most solid tumors employ three main mechanisms to escape immunosurveillance: loss of antigenicity, loss of immunogenicity, and suppressive tumor microenvironment (TME) (51). Sufficient antigenicity is necessary to trigger the initial immune activation. Tumor cells express a diverse population of non-mutated, tumor-specific antigens (TSAs) and tumor-associated antigens (TAAs). TSAs are non-self-antigens only expressed by tumor cells. In contrast, TAAs are self-antigens found in normal cells but upregulated in tumor cells. Ideally, the immune system recognizes these antigens expressed by tumor cells, leading to a potent tumor-specific immune response (52-54). Usually, by selective pressures, tumor cells deprived of TSAs could circumvent immune-mediated attacks to survive and proliferate. There are several proposed mechanisms through which antigenicity could be lost. Immune cells are known to selectively eliminate tumor cells with high tumor mutation burden (TMB), allowing low TMB-tumor cell populations to replicate and thrive (51). Secondly, transformed cells acquire genetic mutations, resulting in MHC-I molecule downregulation and antigen-processing machinery dysfunction, thereby rendering antigen presentation defective in tumor cells (55,56) (Figure 2). In comparison, tumor cells with retained antigenicity may upregulate the inhibitory molecule expression or mask their antigenicity by producing suboptimal neoantigens. Lastly, cancer cells manipulate their TME to make it less favorable for the activation and survival of antitumor immune cells. Loss of immunogenicity and suppressive TME are two complex topics beyond the scope of this review. These two immune evasion mechanisms were discussed in detail in Beatty and Gladney’s review (51).
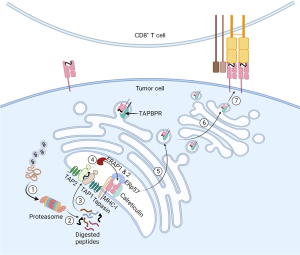
Current efforts in targeting solid tumor antigen presentation
A potential target from the antigen processing and presentation pathway is immunoproteasome. The immunoproteasome was first described in the early 1990s, under the notion that IFN-γ induced changes in the proteasome catalytic subunits, which led to altered catalytic activity (58-62). Furthermore, peptides produced by immunoproteasomes are restricted to the MHC-I antigen processing pathway, which sets it apart from the consecutive proteasome (59,63,64). Due to its specificity only induced by IFN-γ in altered cells, conceptually, immunoproteasome is an optimal drug target for cancer immunotherapy. By selectively targeting the immunoproteasome, the immunopeptidome landscape will show MHC-I preferences, triggering more robust CTL activation and targeted killing. Immunoproteasome deficiency is associated with poor prognosis in various cancer types (65,66). Despite its ostensible advantage, current drugs targeting immunoproteasomes have shown only modest therapeutic efficacy due to the heterogeneous expression of the immunoproteasome in different tumors (67,68). Current research efforts on targeting immunotherapy are listed in Table 1, and other explored targets along the antigen processing and presentation pathway.
Table 1
| Antigen presentation pathway target | Tumor evasion mechanism | Therapeutic approaches | References |
|---|---|---|---|
| Immunoproteasome | Diminished immunopeptidome landscape | LMP7/β5i/PSMB8 inhibitors—ONX-912, PR-924, UK-101, IPSI-001 | (37,70-73) |
| Regulatory subunit stabilizer—ATT-I | |||
| TAP | Reduced antigen: MHC-I complex expression | Increase T cell epitopes associated with impaired peptide processing (TEIPPs) | (74-77) |
| Inhibit TAP with a siRNA vaccine | |||
| TAPBPR | Decreased immunopeptidome repertoire | Introduction of plasma membrane targeted or exogenous soluble TAPBPR | (78-81) |
| ERAP1 & 2 | Altered immunopeptidome | ERAP1 & 2 inhibitors | (82-94) |
LMP7, large multifunctional peptidase 7; PSMB8, proteasome subunit β type 8; ATT-I, atractylenolide-I; TAP, transporter associated with antigen processing; MHC-I, major histocompatibility complex-I; TAPBPR, Tapasin and TAP-binding protein related; ERAP1 & 2, endoplasmic reticulum aminopeptidase 1 & 2.
Therapeutic interventions focused on upregulating tumor antigen presentation in the breast cancer immunotherapy arena have been proposed and investigated. An epigenetic HDAC inhibitor, BML-210, has been studied previously as an effective drug to upregulate MHC-I antigen processing and presentation in triple-negative breast cancer models (TNBC) (95). This study also confirmed the effectiveness of BML-210 and PD-1 mAb as a combination therapy, thus proving that enhanced tumor MHC-I antigen presentation is a rational and effective strategy for designing improved immunotherapeutic. In another study, our laboratory identified MAL2 (Mal, T cell differentiation protein 2) as a crucial player in the recycling of antigen-loaded MHC-I complex in breast cancer models, suggesting MAL2 as a potential novel target for immunotherapy (96). Moreover, a group discovered a long noncoding RNA (lncRNA) LINK-A expression in patients with TNBC. This lncRNA was found to enhance K48-polyubiquitin-mediated degradation of the antigen peptide-load complex, which provided a new foundation for developing combinational immunotherapy treatments (97).
Enhancing antigen presentation to improve immunotherapy has been explored in other cancer types. EZH2 (Enhancer of Zeste 2 polycomb repressive complex 2) was identified as a therapeutic target that enhanced tumor cell antigen presentation in head and neck squamous cell carcinomas (98). NBR1 (Neighbor of Brca1), a cargo receptor responsible for MHC-I degradation through the autophagy-dependent mechanism, was found to be a potential drug target in pancreatic ductal adenocarcinoma (PDAC) (99). An improved analog of indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor, YH29407, has improved T cell infiltration and tumor antigen presentation in murine colorectal carcinoma models (100). Although current studies have demonstrated the therapeutic potential in modulating antigen presentation, more research needs to be done to provide a more robust foundation for treating different tumor types, thereby opening more avenues for personalized medicine.
The association between TCM and immunotherapy (focusing on antigen presentation)
Atractylenolide I (ATT-I) is a sesquiterpene lactone compound found in Atractylodes macrocephala Koidz. Emerging evidence suggests that ATT-I exhibits antitumor effects in various cancer types, including colorectal cancer, melanoma, ovarian cancer, and breast cancer (37,101-104). Recent studies from our group have revealed that binding of natural ATT-I compound with proteasome 26S subunit non-ATPase 4 (PSMD4) stabilizes the immunoproteasome in tumor cells. The stabilization resulted in enhanced MHC-I mediated antigen presentation in tumor cells, thus, triggering CTLs cytotoxicity and killing in the colorectal cancer cell model. Our study also demonstrated enhanced efficacy of ICB therapy by combining ATT-I and PD-1 monoclonal antibody (mAb) in the murine colorectal carcinoma model (37). In breast cancer, ATT-I suppressed tumor growth and metastasis by inhibiting the Toll-like receptor 4 (TLR-4) mediated NF-kB signaling pathway (101). ATT-I has been reported to involve in cell cycle arrest and apoptosis in melanoma and ovarian cancer cells. ATT-I-treated B16 melanoma cells had an increased p21, an indicator for cell cycle arrest, and a decreased CDK2 expression, a key cell cycle progression promoter. Overall, these effects impede cycle cell progression beyond the G1 phase. Furthermore, ATT-I-treated B16 melanoma cells exhibited increased apoptosis activity via increased p53 and decreased ERK/GSK3β signaling (103). In contrast, the effects of ATT-I in A2780 cells, a human ovarian cell line, appeared to be different from that of melanoma cells. ATT-I-treated A2780 cells showed cell cycle arrest in the G2/M phase transition, which was induced by decreased expressions of cyclin B1 and CDK1. In addition, enhanced apoptosis in ATT-I-treated A2780 cells resulted from the altered PI3K/Akt/mTOR pathway (102). An interesting point arose from these studies as the same compound might target multiple cell cycle checkpoints to exert similar inhibitory effects. Thus, more investigations of these pathways and effects on cell cycles by ATT-I should be conducted to delineate whether different mechanisms are involved in distinct types of cancer when treated by ATT-I.
Curcumin is a natural, non-toxic polyphenol substance isolated from the rhizomes of Curcuma longa, Curcuma zedoaria, and Acorus calamus L (105). Supported by extensive studies, curcumin has many pharmacological benefits, such as anti-inflammatory, anti-cancer and immunomodulatory effects (106-111). Many mechanisms of action of curcumin have been proposed and investigated. STAT3 plays a vital role in the receptor tyrosine kinase pathway, which regulates gene transcription. Consecutive activation of STAT3 is observed in many cancer types. Curcumin has been reported to augment in vivo enhancement of CTLs through rejuvenating DCs by directly inhibiting STAT3, a key modulator in the tyrosine kinase pathway with known consecutive activation in many cancer types. In addition, combining curcumin with PD-1/PD-L1 checkpoint blockade agents demonstrated synergistic antitumor effects in murine tumor models (112). In another study, low-dose curcumin revealed an enhanced CTL-mediated antitumor immunity targeting 3LL tumor cells, a Lewis lung carcinoma cell line, by increasing IFN-γ secretion and proliferation of CTLs (108). In head and neck squamous cell carcinoma, curcumin was postulated to downregulate the expression of immune checkpoint (IC) ligands such as PD-L1, PD-L2, and Galectin-9, leading to reduced epithelial-to-mesenchymal transition, restored CTLs cytotoxicity, and reduced CD4+CD25+FoxP3+ Treg cells within the solid TME (109). Despite these promising therapeutic effects, natural curcumin extract has limited utility due to its poor water solubility and stability. A recent study has focused on designing and examining curcumin analogs. One example was GO-Y030, a curcumin analog that limited the immunosuppressive function of Treg cells by inhibiting their mTOR-S6 axis (107).
Recently, more TCM compounds have been investigated in the cancer immunotherapy field. Artesunate, is derived from a natural compound, artemisinin. Artemisinin is extracted from Artemisia annua and is used to treat malaria. Recently, Artesunate has also been identified as a potent anti-cancer candidate. It inhibits TAZ/PD-L1 signaling in non-small cell lung cancer (113). Other small molecule compounds extracted from a TCM called Huangqin have been extensively analyzed in Cai et al.’s in silico study (114). In short, based on their analysis, baicalin, wogonin, and oroxylin A are the bioactive ingredients in Huangqin, which claim to promote anti-tumor immunity. However, these studies have only been preliminary, and more research is needed to validate the efficacy and therapeutic utility of those small molecule compounds.
A general concern about using TCM lies in the poor water solubility, purity, and low efficacy when used in smaller quantities. Thus, analogs of TCM should be explored and assessed to improve the solubility and efficacy of these small molecular compounds to enhance their clinical utility.
Conclusions
It is challenging to treat solid tumors with current immunotherapy strategies due to their complex TME and other factors involved in the targeted killing. However, ample evidence has suggested that enhanced antigen presentation by the tumor cells can be a promising way to induce cytotoxicity by the CD8+ effector T cells, thereby leading to tumor shrinkage and clinical remission. The precise antigen processing and presentation mechanism in cancer cells are still unclear. A deeper understanding into such pathways would offer more insight into the heterogeneous nature of the antigenicity and immunogenicity of tumor cells. Furthermore, more research is needed to figure out if there are possible small molecule inhibitors that could potentiate the MHC-I antigen presentation pathway in tumor cells to enhance the cytotoxicity of CTLs and accelerate tumor apoptosis.
Acknowledgments
Figures are created with BioRender.com.
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://lcm.amegroups.com/article/view/10.21037/lcm-23-5/coif). XL has a pending international patent application No. PCT/US2021/061295, entitled “Methods to sensitize cancer cells to immune attack using atractylenolide”. XL serves as an unpaid editorial board member of Longhua Chinese Medicine from September 2022 to August 2024. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schnell A, Bod L, Madi A, et al. The yin and yang of co-inhibitory receptors: toward anti-tumor immunity without autoimmunity. Cell Res 2020;30:285-99. [Crossref] [PubMed]
- Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta 2009;1790:963-9. [Crossref] [PubMed]
- Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science 2017;357:eaal2380. [Crossref] [PubMed]
- Grønbaek K, Treppendahl M, Asmar F, et al. Epigenetic changes in cancer as potential targets for prophylaxis and maintenance therapy. Basic Clin Pharmacol Toxicol 2008;103:389-96. [Crossref] [PubMed]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007;26:7773-9. [Crossref] [PubMed]
- Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene 2008;27:2801-9. [Crossref] [PubMed]
- Felsher DW. Oncogene addiction versus oncogene amnesia: perhaps more than just a bad habit? Cancer Res 2008;68:3081-6; discussion 3086. [Crossref] [PubMed]
- Shortt J, Johnstone RW. Oncogenes in cell survival and cell death. Cold Spring Harb Perspect Biol 2012;4:a009829. [Crossref] [PubMed]
- Warrington R, Watson W, Kim HL, et al. An introduction to immunology and immunopathology. Allergy Asthma Clin Immunol 2011;7:S1. [Crossref] [PubMed]
- Cheng M, Chen Y, Xiao W, et al. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol 2013;10:230-52. [Crossref] [PubMed]
- Taylor BC, Balko JM. Mechanisms of MHC-I Downregulation and Role in Immunotherapy Response. Front Immunol 2022;13:844866. [Crossref] [PubMed]
- Montesion M, Murugesan K, Jin DX, et al. Somatic HLA Class I Loss Is a Widespread Mechanism of Immune Evasion Which Refines the Use of Tumor Mutational Burden as a Biomarker of Checkpoint Inhibitor Response. Cancer Discov 2021;11:282-92. [Crossref] [PubMed]
- Cornel AM, Mimpen IL, Nierkens S. MHC Class I Downregulation in Cancer: Underlying Mechanisms and Potential Targets for Cancer Immunotherapy. Cancers (Basel) 2020;12:1760. [Crossref] [PubMed]
- Vivier E, Tomasello E, Baratin M, et al. Functions of natural killer cells. Nat Immunol 2008;9:503-10. [Crossref] [PubMed]
- Murphy K, Weaver C. Janeway’s Immunobiology. 9th edition. New York, NY: Garland Science/Taylor & Francis Group. 2017.
- Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol 2020;17:807-21. [Crossref] [PubMed]
- Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol 2020;20:651-68. [Crossref] [PubMed]
- Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol 2002;2:309-22. [Crossref] [PubMed]
- Jenkins MK, Chu HH, McLachlan JB, et al. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol 2010;28:275-94. [Crossref] [PubMed]
- Wölfl M, Greenberg PD. Antigen-specific activation and cytokine-facilitated expansion of naive, human CD8+ T cells. Nat Protoc 2014;9:950-66. [Crossref] [PubMed]
- Curtsinger JM, Mescher MF. Inflammatory cytokines as a third signal for T cell activation. Curr Opin Immunol 2010;22:333-40. [Crossref] [PubMed]
- Boon T, Cerottini JC, Van den Eynde B, et al. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol 1994;12:337-65. [Crossref] [PubMed]
- Rha MS, Shin EC. Activation or exhaustion of CD8+ T cells in patients with COVID-19. Cell Mol Immunol 2021;18:2325-33. [Crossref] [PubMed]
- Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486-99. [Crossref] [PubMed]
- Brooks DG, McGavern DB, Oldstone MB. Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J Clin Invest 2006;116:1675-85. [Crossref] [PubMed]
- Wherry EJ, Blattman JN, Murali-Krishna K, et al. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 2003;77:4911-27. [Crossref] [PubMed]
- Thommen DS, Schumacher TN. T Cell Dysfunction in Cancer. Cancer Cell 2018;33:547-62. [Crossref] [PubMed]
- McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 2019;37:457-95. [Crossref] [PubMed]
- Gumber D, Wang LD. Improving CAR-T immunotherapy: Overcoming the challenges of T cell exhaustion. EBioMedicine 2022;77:103941. [Crossref] [PubMed]
- Raskov H, Orhan A, Christensen JP, et al. Cytotoxic CD8(+) T cells in cancer and cancer immunotherapy. Br J Cancer 2021;124:359-67. [Crossref] [PubMed]
- Durgeau A, Virk Y, Corgnac S, et al. Recent Advances in Targeting CD8 T-Cell Immunity for More Effective Cancer Immunotherapy. Front Immunol 2018;9:14. [Crossref] [PubMed]
- Jackson SR, Yuan J, Teague RM. Targeting CD8+ T-cell tolerance for cancer immunotherapy. Immunotherapy 2014;6:833-52. [Crossref] [PubMed]
- Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol 2019;234:8509-21. [Crossref] [PubMed]
- Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nat Rev Immunol 2018;18:527-35. [Crossref] [PubMed]
- Weigelin B, Friedl P. T cell-mediated additive cytotoxicity - death by multiple bullets. Trends Cancer 2022;8:980-7. [Crossref] [PubMed]
- Weigelin B, den Boer AT, Wagena E, et al. Cytotoxic T cells are able to efficiently eliminate cancer cells by additive cytotoxicity. Nat Commun 2021;12:5217. [Crossref] [PubMed]
- Xu H, Van der Jeught K, Zhou Z, et al. Atractylenolide I enhances responsiveness to immune checkpoint blockade therapy by activating tumor antigen presentation. J Clin Invest 2021;131:e146832. [Crossref] [PubMed]
- Jorgovanovic D, Song M, Wang L, et al. Roles of IFN-γ in tumor progression and regression: a review. Biomark Res 2020;8:49. [Crossref] [PubMed]
- Mo X, Zhang H, Preston S, et al. Interferon-γ Signaling in Melanocytes and Melanoma Cells Regulates Expression of CTLA-4. Cancer Res 2018;78:436-50. [Crossref] [PubMed]
- Ventola CL. Cancer Immunotherapy, Part 1: Current Strategies and Agents. P T 2017;42:375-83. [PubMed]
- Zhu S, Zhang T, Zheng L, et al. Combination strategies to maximize the benefits of cancer immunotherapy. J Hematol Oncol 2021;14:156. [Crossref] [PubMed]
- Zhao B, Zhao H, Zhao J. Efficacy of PD-1/PD-L1 blockade monotherapy in clinical trials. Ther Adv Med Oncol 2020;12:1758835920937612. [Crossref] [PubMed]
- Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711-23. [Crossref] [PubMed]
- Paz-Ares LG, Ramalingam SS, Ciuleanu TE, et al. First-Line Nivolumab Plus Ipilimumab in Advanced NSCLC: 4-Year Outcomes From the Randomized, Open-Label, Phase 3 CheckMate 227 Part 1 Trial. J Thorac Oncol 2022;17:289-308. [Crossref] [PubMed]
- Reck M, Remon J, Hellmann MD. First-Line Immunotherapy for Non-Small-Cell Lung Cancer. J Clin Oncol 2022;40:586-97. [Crossref] [PubMed]
- Shields MD, Marin-Acevedo JA, Pellini B. Immunotherapy for Advanced Non-Small Cell Lung Cancer: A Decade of Progress. Am Soc Clin Oncol Educ Book 2021;41:1-23. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med 2019;381:1535-46. [Crossref] [PubMed]
- Schadendorf D, Hodi FS, Robert C, et al. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol 2015;33:1889-94. [Crossref] [PubMed]
- Morinaga T, Inozume T, Kawazu M, et al. Mixed Response to Cancer Immunotherapy is Driven by Intratumor Heterogeneity and Differential Interlesion Immune Infiltration. Cancer Res Commun 2022;2:739-53. [Crossref] [PubMed]
- Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res 2015;21:687-92. [Crossref] [PubMed]
- Leko V, Rosenberg SA. Identifying and Targeting Human Tumor Antigens for T Cell-Based Immunotherapy of Solid Tumors. Cancer Cell 2020;38:454-72. [Crossref] [PubMed]
- Wagner S, Mullins CS, Linnebacher M. Colorectal cancer vaccines: Tumor-associated antigens vs neoantigens. World J Gastroenterol 2018;24:5418-32. [Crossref] [PubMed]
- Zhang Z, Lu M, Qin Y, et al. Neoantigen: A New Breakthrough in Tumor Immunotherapy. Front Immunol 2021;12:672356. [Crossref] [PubMed]
- Bandola-Simon J, Roche PA. Dysfunction of antigen processing and presentation by dendritic cells in cancer. Mol Immunol 2019;113:31-7. [Crossref] [PubMed]
- Dhatchinamoorthy K, Colbert JD, Rock KL. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front Immunol 2021;12:636568. [Crossref] [PubMed]
- Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol 2008;8:607-18. [Crossref] [PubMed]
- Tripathi SC, Vedpathak D, Ostrin EJ. The Functional and Mechanistic Roles of Immunoproteasome Subunits in Cancer. Cells 2021;10:3587. [Crossref] [PubMed]
- Goldberg AL, Cascio P, Saric T, et al. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol 2002;39:147-64. [Crossref] [PubMed]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 1999;68:1015-68. [Crossref] [PubMed]
- Collins GA, Goldberg AL. The Logic of the 26S Proteasome. Cell 2017;169:792-806. [Crossref] [PubMed]
- Aki M, Shimbara N, Takashina M, et al. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem 1994;115:257-69. [Crossref] [PubMed]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol 1999;17:739-79. [Crossref] [PubMed]
- Rock KL, Gramm C, Rothstein L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994;78:761-71. [Crossref] [PubMed]
- Kalaora S, Lee JS, Barnea E, et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat Commun 2020;11:896. [Crossref] [PubMed]
- Tripathi SC, Peters HL, Taguchi A, et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc Natl Acad Sci U S A 2016;113:E1555-64. [Crossref] [PubMed]
- Singh AV, Bandi M, Aujay MA, et al. PR-924, a selective inhibitor of the immunoproteasome subunit LMP-7, blocks multiple myeloma cell growth both in vitro and in vivo. Br J Haematol 2011;152:155-63. [Crossref] [PubMed]
- Xu H, Ju D, Jarois T, et al. Diminished feedback regulation of proteasome expression and resistance to proteasome inhibitors in breast cancer cells. Breast Cancer Res Treat 2008;107:267-74. [Crossref] [PubMed]
- D'Amico S, Tempora P, Melaiu O, et al. Targeting the antigen processing and presentation pathway to overcome resistance to immune checkpoint therapy. Front Immunol 2022;13:948297. [Crossref] [PubMed]
- Vachharajani N, Joeris T, Luu M, et al. Prevention of colitis-associated cancer by selective targeting of immunoproteasome subunit LMP7. Oncotarget 2017;8:50447-59. [Crossref] [PubMed]
- Koerner J, Brunner T, Groettrup M. Inhibition and deficiency of the immunoproteasome subunit LMP7 suppress the development and progression of colorectal carcinoma in mice. Oncotarget 2017;8:50873-88. [Crossref] [PubMed]
- Wehenkel M, Ban JO, Ho YK, et al. A selective inhibitor of the immunoproteasome subunit LMP2 induces apoptosis in PC-3 cells and suppresses tumour growth in nude mice. Br J Cancer 2012;107:53-62. [Crossref] [PubMed]
- Kuhn DJ, Hunsucker SA, Chen Q, et al. Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood 2009;113:4667-76. [Crossref] [PubMed]
- Durgeau A, Virk Y, Gros G, et al. Human preprocalcitonin self-antigen generates TAP-dependent and -independent epitopes triggering optimised T-cell responses toward immune-escaped tumours. Nat Commun 2018;9:5097. [Crossref] [PubMed]
- Garrido G, Schrand B, Rabasa A, et al. Tumor-targeted silencing of the peptide transporter TAP induces potent antitumor immunity. Nat Commun 2019;10:3773. [Crossref] [PubMed]
- Doorduijn EM, Sluijter M, Querido BJ, et al. TAP-independent self-peptides enhance T cell recognition of immune-escaped tumors. J Clin Invest 2016;126:784-94. [Crossref] [PubMed]
- Marijt KA, Blijleven L, Verdegaal EME, et al. Identification of non-mutated neoantigens presented by TAP-deficient tumors. J Exp Med 2018;215:2325-37. [Crossref] [PubMed]
- Hermann C, Trowsdale J, Boyle LH. TAPBPR: a new player in the MHC class I presentation pathway. Tissue Antigens 2015;85:155-66. [Crossref] [PubMed]
- Hafstrand I, Aflalo A, Boyle LH. Why TAPBPR? Implications of an additional player in MHC class I peptide presentation. Curr Opin Immunol 2021;70:90-4. [Crossref] [PubMed]
- Ilca FT, Drexhage LZ, Brewin G, et al. Distinct Polymorphisms in HLA Class I Molecules Govern Their Susceptibility to Peptide Editing by TAPBPR. Cell Rep 2019;29:1621-1632.e3. [Crossref] [PubMed]
- Ilca FT, Neerincx A, Wills MR, et al. Utilizing TAPBPR to promote exogenous peptide loading onto cell surface MHC I molecules. Proc Natl Acad Sci U S A 2018;115:E9353-61. [Crossref] [PubMed]
- Temponeras I, Stamatakis G, Samiotaki M, et al. ERAP2 Inhibition Induces Cell-Surface Presentation by MOLT-4 Leukemia Cancer Cells of Many Novel and Potentially Antigenic Peptides. Int J Mol Sci 2022;23:1913. [Crossref] [PubMed]
- López de Castro JA. How ERAP1 and ERAP2 Shape the Peptidomes of Disease-Associated MHC-I Proteins. Front Immunol 2018;9:2463. [Crossref] [PubMed]
- Koumantou D, Barnea E, Martin-Esteban A, et al. Editing the immunopeptidome of melanoma cells using a potent inhibitor of endoplasmic reticulum aminopeptidase 1 (ERAP1). Cancer Immunol Immunother 2019;68:1245-61. [Crossref] [PubMed]
- Textoris-Taube K, Cammann C, Henklein P, et al. ER-aminopeptidase 1 determines the processing and presentation of an immunotherapy-relevant melanoma epitope. Eur J Immunol 2020;50:270-83. [Crossref] [PubMed]
- Keller M, Ebstein F, Bürger E, et al. The proteasome immunosubunits, PA28 and ER-aminopeptidase 1 protect melanoma cells from efficient MART-126-35 -specific T-cell recognition. Eur J Immunol 2015;45:3257-68. [Crossref] [PubMed]
- James E, Bailey I, Sugiyarto G, et al. Induction of protective antitumor immunity through attenuation of ERAAP function. J Immunol 2013;190:5839-46. [Crossref] [PubMed]
- D'Amico S, D'Alicandro V, Compagnone M, et al. ERAP1 Controls the Interaction of the Inhibitory Receptor KIR3DL1 With HLA-B51:01 by Affecting Natural Killer Cell Function. Front Immunol 2021;12:778103. [Crossref] [PubMed]
- Cifaldi L, Romania P, Falco M, et al. ERAP1 regulates natural killer cell function by controlling the engagement of inhibitory receptors. Cancer Res 2015;75:824-34. [Crossref] [PubMed]
- Cifaldi L, Lo Monaco E, Forloni M, et al. Natural killer cells efficiently reject lymphoma silenced for the endoplasmic reticulum aminopeptidase associated with antigen processing. Cancer Res 2011;71:1597-606. [Crossref] [PubMed]
- Firat E, Saveanu L, Aichele P, et al. The role of endoplasmic reticulum-associated aminopeptidase 1 in immunity to infection and in cross-presentation. J Immunol 2007;178:2241-8. [Crossref] [PubMed]
- York IA, Brehm MA, Zendzian S, et al. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A 2006;103:9202-7. [Crossref] [PubMed]
- Yan J, Parekh VV, Mendez-Fernandez Y, et al. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J Exp Med 2006;203:647-59. [Crossref] [PubMed]
- Hammer GE, Gonzalez F, James E, et al. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol 2007;8:101-8. [Crossref] [PubMed]
- Zhou Z, Van der Jeught K, Fang Y, et al. An organoid-based screen for epigenetic inhibitors that stimulate antigen presentation and potentiate T-cell-mediated cytotoxicity. Nat Biomed Eng 2021;5:1320-35. [Crossref] [PubMed]
- Fang Y, Wang L, Wan C, et al. MAL2 drives immune evasion in breast cancer by suppressing tumor antigen presentation. J Clin Invest 2021;131:e140837. [Crossref] [PubMed]
- Hu Q, Ye Y, Chan LC, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol 2019;20:835-51. [Crossref] [PubMed]
- Zhou L, Mudianto T, Ma X, et al. Targeting EZH2 Enhances Antigen Presentation, Antitumor Immunity, and Circumvents Anti-PD-1 Resistance in Head and Neck Cancer. Clin Cancer Res 2020;26:290-300. [Crossref] [PubMed]
- Yamamoto K, Venida A, Yano J, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020;581:100-5. [Crossref] [PubMed]
- Kim DK, Synn CB, Yang SM, et al. YH29407 with anti-PD-1 ameliorates anti-tumor effects via increased T cell functionality and antigen presenting machinery in the tumor microenvironment. Front Chem 2022;10:998013. [Crossref] [PubMed]
- Long F, Lin H, Zhang X, et al. Atractylenolide-I Suppresses Tumorigenesis of Breast Cancer by Inhibiting Toll-Like Receptor 4-Mediated Nuclear Factor-κB Signaling Pathway. Front Pharmacol 2020;11:598939. [Crossref] [PubMed]
- Long F, Wang T, Jia P, et al. Anti-Tumor Effects of Atractylenolide-I on Human Ovarian Cancer Cells. Med Sci Monit 2017;23:571-9. [Crossref] [PubMed]
- Ye Y, Chao XJ, Wu JF, et al. ERK/GSK3β signaling is involved in atractylenolide I-induced apoptosis and cell cycle arrest in melanoma cells. Oncol Rep 2015;34:1543-8. [Crossref] [PubMed]
- Sun Y, Liu Y, Cai Y, et al. Atractylenolide I inhibited the development of malignant colorectal cancer cells and enhanced oxaliplatin sensitivity through the PDK1-FoxO1 axis. J Gastrointest Oncol 2022;13:2382-92. [Crossref] [PubMed]
- Deng LJ, Qi M, Li N, et al. Natural products and their derivatives: Promising modulators of tumor immunotherapy. J Leukoc Biol 2020;108:493-508. [Crossref] [PubMed]
- Shafabakhsh R, Pourhanifeh MH, Mirzaei HR, et al. Targeting regulatory T cells by curcumin: A potential for cancer immunotherapy. Pharmacol Res 2019;147:104353. [Crossref] [PubMed]
- MaruYama T. Curcumin analog GO-Y030 boosts the efficacy of anti-PD-1 cancer immunotherapy. Cancer Sci 2021;112:4844-52. [Crossref] [PubMed]
- Luo F, Song X, Zhang Y, et al. Low-dose curcumin leads to the inhibition of tumor growth via enhancing CTL-mediated antitumor immunity. Int Immunopharmacol 2011;11:1234-40. [Crossref] [PubMed]
- Liu L, Lim MA, Jung SN, et al. The effect of Curcumin on multi-level immune checkpoint blockade and T cell dysfunction in head and neck cancer. Phytomedicine 2021;92:153758. [Crossref] [PubMed]
- Hayakawa T, Yaguchi T, Kawakami Y. Enhanced anti-tumor effects of the PD-1 blockade combined with a highly absorptive form of curcumin targeting STAT3. Cancer Sci 2020;111:4326-35. [Crossref] [PubMed]
- Luo H, Vong CT, Chen H, et al. Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine. Chin Med 2019;14:48. [Crossref] [PubMed]
- Huynh J, Chand A, Gough D, et al. Therapeutically exploiting STAT3 activity in cancer - using tissue repair as a road map. Nat Rev Cancer 2019;19:82-96. [Crossref] [PubMed]
- Cao D, Chen D, Xia JN, et al. Artesunate promoted anti-tumor immunity and overcame EGFR-TKI resistance in non-small-cell lung cancer by enhancing oncogenic TAZ degradation. Biomed Pharmacother 2022;155:113705. [Crossref] [PubMed]
- Cai C, Wu Q, Hong H, et al. In silico identification of natural products from Traditional Chinese Medicine for cancer immunotherapy. Sci Rep 2021;11:3332. [Crossref] [PubMed]
Cite this article as: Wang X, Lu X. Modern transformation of ancient healing arts: traditional Chinese medicine in cancer immunotherapy. Longhua Chin Med 2023;6:1.

