
Narrative review of the progress in discovery of anti COVID-19 agents from traditional Chinese medicine using in silico approaches
Introduction
A major threat to global health is ongoing outbreak of the respiratory disease named COVID-19 (1) that was declared pandemic by the World Health Organization when the number of confirmed cases approached 200,000 with over 8,000 deaths (2). Currently, October 2020, there are over 33 million confirmed cases and over 1 million deaths. The COVID-19 is causing enormous challenges not only for public health and medical communities, but also for economy (3), society (4) and mental health (5).
COVID-19 is caused by highly virulent SARS-CoV-2 virus that belongs to the Betacoronavirus genus. The rapid spread of the disease, with no or very little knowledge about effective treatments, instigated hunt for COVID-19 treatments and engaged research groups to identify effective anti-viral agents or to develop a vaccine. As a part of these efforts, structural studies provided three-dimensional (3D) structures of viral target proteins such as main protease (Mpro) (6) and spike (S) protein (7) amongst others. Such knowledge provide a basis for the structure-based drug discovery (SBDD) (8) and drug repurposing (9). The extensive reviews on drug targets and early efforts in the discovery of the agents for the COVID-19 treatment are given elsewhere (10-12).
The aim of this mini review is to explore the use of computational chemistry to enhance the efforts to utilize traditional Chinese medicine (TCM) of the COVID-19 disease. The focus of this review is on the identification of potential anti-viral agents from TCM herbs or formulations and/or to find rationale for their use in the clinic by conducting in silico mechanistic studies. This review will cover selected case studies and it is not a comprehensive review of all literature published in the field.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/lcm-20-42).
Methods
Data sources
The Google Scholar and Scopus databases were used to perform this review (from February 2020 to August 2020).
Focal question
This review was carried out to explore the use of in silico methods to evaluate potential Chinese medicine for the treatment of COVID-19.
Search
The searches of the databases were conducted using a combination of keywords “traditional Chinese medicine” “COVID-19” with one of the following keywords “in silico”, “docking”, “molecular dynamics simulation” and “virtual screening”.
Results and discussion
Virtual screening of components extracted from Chinese herbal medicine (CHM) plants
Flavonoids, often found in food and in CHM plants, were investigated for their potential to favourably interact with SARS-CoV-2 proteins and disrupt its replication. AutoDock Vina was used to conduct the docking of 23 natural flavonoids against three targets, amin protease (Mpro), RNA dependent RNA polymerase (rdrp) and spike (S) protein. The 3D structures of these proteins were not available and homology modelling was conducted using SwissModel online server using PDB entries 6M71, 6YB7 and 6LZG as template structures. All natural products have shown the propensity to interact with these three proteins at their active surfaces. However, it was predicted that quercetin 1, a compounds with an established anti-viral activity, has a greatest potential to establish favourable interactions with Mpro at Glu290 and Asp289, as well as with the receptor binding domain of the viral spike (13).
Mpro was selected for a different docking study, but protein target structure was extracted from PDB entry 6LU7, a crystal structure of the Mpro with an inhibitor N3 (14). The set of natural products was established based on their previously established anti-viral activity against different viruses, including SARS-Coronavirus and were downloaded from the PubChem Database. The compounds from the set that are found in TCM plants were glycyrrhizin (CID:128229), indirubin (CID:10177), indican (CID:441564), indigo (CID:10215), hesperetin (CID:72281) The docking was conducted using AutoDock Vina (15). Amongst those, it was found that glycyrrhizin have most favourable interaction with the target protein by forming five hydrogen bonds with residues from the protease active site (GLN 127, LYS 5, LYS 137, ARG 131 and TYR 239). Despite predicted favourable interactions, glycyrrhizin is not a drug-like molecule as it violates three of Lipinski’s rule of five. Therefore, this molecule can provide a basis for a further development of lead compound against COVID-19.
In silico investigation of a potential for anti-COVID-19 activity was conducted on the selected chemical class, saikosaponins found in TCM plants such as Bupleurum spp., Heteromorpha spp. and Scrophularia scorodonia (16,17). A set of 23 molecules was used in a docking procedure against NSP15 endoribonuclease from SARS-CoV-2, PDB entry 6W01, (18) and prefusion 2019-nCoV spike glycoprotein, PDB entry 6VSB (7). Docking using GLIDE software revealed that the saikosaponins U and V have potential for favourable interactions with both proteins. Such interactions could potentially both affect the virus replication and curtail the cell entry (19).
Molecular docking was used to explore interactions of components of CHM with two selected targets ACE2 and SARS-Cov-2 3CL (20). Target protein structures were downloaded as PDB entries 1R42 (21) and 6LU7 (14), respectively, and used without further modifications. A total of 12,541 components of CHM were downloaded from a TCM systems pharmacology database and analysis platform (TCMSP) (22). These components were docked against both targets using Autodock Vina (15). Top six hits were TCM components (puerarin, bicuculline, luteolin, quercetin and isorhamnetin) and had a better docking score than clinical drugs (lopinavir, ritonavir and remdesivir). These compounds can be found in 238 CHM that could have potential anti-COVID-19 effect, with Forsythiae fructus and Lonicerae Japonicae flos as CHM with most containing possible active ingredients. Network pharmacology also revealed that these two plants could act on 121 targets and 14 biochemical pathways related to viral pneumonia (20), and therefore, could potentially be used in the treatment of COVID-19 in combination with Western medicine. The multi-component and multi-target aspects of CHM may play considerable roles in anti-inflammatory and immunomodulatory response to viral infection.
The other approach was to explore the potential of TCM components to prevent host-pathogen interaction by targeting the protein-protein interaction (PPI) surface between human angiotensin-converting enzyme 2 (ACE2) and SARS-Cov-2 spike protein. One of the approaches was to select potentially active compounds based on the anecdotal evidence of their use in the treatment of COVID-19. Caflanone, hesperetin and myricetin are known to have anti-viral activity (23) and were chosen to establish their affinity for ACE2 enzyme (24). Induced fit docking was conducted using Schrodinger suite and it was found that these flavonoids have affinity for the sites different than the binding site of chloroquine (25), a molecule currently in the clinical trials for the treatment of COVID-19 infections. These promising results in silico studies instigated in vitro studies that suggest that caflanone has a potential to inhibit virus entry factors including cathepsin L as well as cytokines that may be relevant to response of human macrophages to infection with SARS-CoV.
Astragaloside IV (PubChem, CID13943297) is the major active compound amongst 40 compounds isolated from Astragalus mongholicus root that exerts anti-inflammatory effects. This compound is found in a well-known Chinese tonic, Huangqi (Radix Astragali Mongholici) that is used by patients with kidney disease, viral hepatitis and cardiovascular conditions. Over 400 potential targets were identified using SwissTargetPrediction, Comparative Toxicogenomics Database (26), TargetNet (27) and PharmMapper (28). The list of these predicted targets was used to obtain pathways that could be affected by astragaloside IV using Metascape (29). Additionally, protein-protein association network that is involved in COVID-19 was predicted using the knowledge of differentially expressed genes as a result of SARS-CoV-2 infections and STRING webserver (30). The overlap between targets involved in COVID-19 those that predicted for astragaloside IV revealed that 12 proteins are common including NF-κB-p65 and IL-6. Although the docking scores, obtained by Autodock Vina, of this natural product against all 12 targets are generally lower when compared to the docking scores of rapamycin, it is still possible that astragaloside IV may potentially alleviate the effects of the cytokine storm in the lungs that is caused by COVID-19. Additionally, there may be additional beneficial effects due to the multi-level and multi-target pharmacological responses in the use of astragaloside IV in the treatment of COVID-19 (31).
Peach [Prunus persica (L.) Batsch] was recently recognized in the TCM as a contributor to the prevention and treatment of COVID-19 (32). The isolation and identification of potential active components has revealed a novel compound (3'-hydroxy-4'-methoxy-chroman-7-O-β-D-glucopyranoside) and three already known molecules (ferulic acid heptyl ester, naringenin and 4,2',4'-trihydroxy-6'-methoxychalcone-4'-O-β-D-glucopyranoside). Their structures were used for in silico evaluation of their potential to combat COVID-19. Their structures were used for the docking against two viral targets, evaluating their molecular similarity to know anti-viral drugs and prediction of their ADMT properties. Two structures of Mpro were extracted from PDB entries 6LU7 (14) and 6Y2F (6), while the structure of spike protein was extracted from 6VSB (7). Glucopyranoside containing molecules were shown to interact favourably with both target proteins. Additionally, molecular similarity determined using ROCS software (33) indicated that these molecules have likeness to drugs that could prevent either entry of SARS-CoV into lung cells or inflammatory response that could cause lung injury. Therefore, it was suggested that these two glucopyranoside compounds may be candidates for drug development.
There are similar studies that did not go through the peer review process. Preliminary report on the docking study of natural products into ACE2 protein target (PDB entry not defined) indicated that four molecules, baicalin (Scutellaria baicalensis georgi), scutellarin (Erigeron breviscapus), hesperetin (Citri reticulatae pericarpium) and glycyrrhizin (Glycyrrhiza radix), have a potential to exert anti-viral effects and possibly prevent COVID-19 infection (34). Rutin, a component of Lianhua Qingwen (LHQW), is known to inhibit different proteases, including viral Mpro. The mechanism of the action of rutin was explored by molecular docking using Autodock Vina followed by molecular dynamics simulation using NAMD (35). Although it was revealed that rutin fits into Mpro pocket, the binding is not stable due to its hydrophilicity and the part of the molecule leaves the active site during molecular dynamics simulation. This suggests that rutin molecule would not be an optimal inhibitor, but it could provide a basis for the design of more active analogues (36).
Network pharmacology and target identification for TCM decoctions and formulations
Decoctions are more complex mixtures of natural products obtained by boiling water extractions of components from plants used in TCM. They are often used in treatment of viral infections and as in the case of many of the formulations, some of those were used in the COVID-19 treatment based on the anecdotal evidence.
Multicomponent formula Qingfei Paidu decoction (QFPD) was recommended as a possible treatment of COVID-19 in February 2020, however the mechanism of action of this formulation is elusive. The composition of five key herbal components were extracted from the TCMSP with 175 effective compounds selected for further study. The prediction of target genes were predicted using BATMAN-TCM tool (37) and revealed 300 of potential QFPD targets. The COVID-19 docking server (38) revealed that nine compounds with favourable ADMET properties can favourably interact with 12 COVID-19 targets including Mpro. Network analysis indicated that QFPD containing those molecules (PubChem IDs: CID6918970, CID3902, CID14135323, CID42607889, CID440833, CID177149, CID11438306, CID712316 and CID373261) can potentially aid the regulation of metabolic and pro-inflammatory processes to combat infection by SARS-Cov-2 virus (39).
Two decoctions, QFPD and Ma Xing Shi Gan decoction (MXSGD), were characterized using ultraperformance liquid chromatography (UPLC) by identifying compounds in the extracts (40). The therapeutic potential of these decoctions was assessed by querying databases of potential targets, namely the encyclopaedia of traditional Chinese medicine (ETCM) (41), SymMap (42), SwissTargetPrediction (43) and David (44) by establishing their therapeutic networks. The COVID-19 disease network was established using a limited data on the symptoms and known treatment options at the time (March 2020) and querying SymMap database followed by target enrichment analysis by using DAVID database. These results, visualized using Cytoscape (45) and Power BI (46), allowed overlapping the targets identified for the decoctions components with the COVID-19 disease network. It was found QFP could potentially affect 24 target genes in the Toll-like signalling pathway, with three of those are related to drug therapy. Additionally, it was experimentally demonstrated that MXSGD has anti-platelet aggregation effect, and it can be used in conjunction with QFPD to achieve synergistic effects in the treatment of COVID-19 due to change of the content of their components, ephedrine and glycyrrhizic acid (40).
The lung-cleaning and toxicity-excluding (LCTE) soup, Chinese herbal formula, has been used in treating COVID-19 infections, however, without a rationale in terms of TCM. Therefore, all compounds from twenty plants used in the formulation were identified using TCMSP, ECTM and SymMap, and only molecules that had favourable ADME properties for oral delivery were used in the network analysis. DisGeNet (47) database was used to identify protein targets related to the key COVID-19 symptoms. Pathways of the plants-compound-symptom network were constructed using igraph R package and visualized using Cytoscape, while potential in vivo effects were predicted by KEGG pathway (48,49) and Disease Ontology enrichment (50) databases by mapping all proteins that can be targeted by the compounds founds in LCTE soup. Analysis of herb-compound-protein-symptom networks found that mainly six compounds from LCTE (quercetin, kaempferol, luteolin, beta-sitosterol, stigmasterol and naringenin) may act on tumour necrosis factor (TNF) and Solute Carrier Family 6 Member 4 (SLC6A4) as those contribute to alleviating more than five COVID-19 symptoms. Additionally, it was shown that LCTE compounds may be affecting 30 enriched KEGG pathways related to infections by viruses and other microorganisms, as well as the pathways related to pneumonia caused by coronavirus. This study provides a strong rationale of the LCTE soup effectiveness in alleviating main symptoms of COVID-19, while it also alerts to possible effects due to off-target interactions and compound-compound interactions (51).
Similar approaches, where network pharmacology combined with the molecular docking were used to rationalize the mechanism of action for other TCM formulations, such as Qing-Fei-Da-Yuan (QFDY) granules (52) and Cold-Damp Plague (CDP) formula (53) amongst others. The constituents were identified from TCMSP database in both cases, and although the methods for prediction of their potential targets were different, in both cases STRING and KEGG database were used to propose PPI network with relevant pathways and propose most likely targets for the formulation constituents. The docking results obtained by Discovery Studio showed that quercetin, luteolin and naringenin favourably interact with the COVID-19 3CL hydrolase, while anemarsaponin and medicocarpin have a potential to interact with ACE2. The constituents of CDP formulation were docked using Autodock Vina against IL-6 and ACE2 as targets. It was found that highest potential to be most active were quercetin and luteolin against IL-6, while cl-tyrosine and dl-phenylalanine were interacting most favourably with ACE2. Both formulations have a potential to regulate multiple signalling pathways via synergistic effects of herbal components. Summary of other studies based on the network pharmacology combined with molecular docking is shown in Table 1.

Full table
Homology modelling and molecular dynamics
Despite the rapid determination of 3D structures of relevant druggable SARS-CoV-2 virus proteins, there are some proteins that are attractive targets but without experimentally determined structures such as guanine-N7 methyltransferase. This protein is responsible for cloaking the 5'-ends of viral genomic RNA from hosts’ innate immunity and therefore offers opportunities for drug development. Homology model of this protein was generated based on the structure of SARS-CoV nsp14-nsp10 protein (PDB entry: 5C8S) as a template and it was used for a virtual screening for the potential inhibitors. Five molecules from the set of TCM compounds (TCM ID: 57025, 3495, 31007, 20111 nad 5376) were identified as good binders into the active site. Molecular dynamics simulations of all protein-TCM compounds showed that all complexes have adequate stability of complexes and dynamic behaviour, indicating that these five phytochemicals may have strong potential to be used as anti-viral agents (60).
Molecular dynamics simulations is indispensable method in the SBDD (61), however, it is computationally more demanding than molecular docking and virtual screening. Therefore, due to the urgency to respond to quick spreading of the COVID-19 and lack of the effective therapies, there was limited use of molecular dynamics simulations in studies that involved TCM. Nevertheless, the advantages that molecular dynamics simulation methods offer will be needed to make further progress in prediction of TCM components interaction with desired targets, their off-target interactions as well as component-component interactions in such complex formulations.
Conclusions
This mini review demonstrates that scientific community has responded quickly in to address challenges due to the lack of the COVID-19 treatments. Molecular docking and virtual screening were adopted as most pragmatic and least time-consuming approaches that were complementing timely release of structural data for the target proteins. Additionally, due to the complexity of the TCM formulations many studies were employing network pharmacology approach to understand potential synergistic effects due to their possible actions on multiple targets within host and virus.
It was already recognized that the TCM databases are being used for the rapid assessment of the potential of TCM to protect against SARS-CoV-2 infections and/or treat COVID-19 disease. Additional, these databases are also used theoretical mechanistic studies of TCM against COVID-19 and may provide rationale for the action of remedies used in clinic (62). However, these database-based studies are mainly theoretical and conducted over the short period that did not allow experimental validation of the theoretical studies. In 80 studies that were reported until mid-June 2020, 36 TCM formulations were not validated experimentally for the use for in clinic. Due to the lack of information on component interactions and off-target interactions, such studies may overestimate the potential effects of these TCMs in the treatment of COVID-19 (63).
It is notable that some TCM components (Figure 1) are identified as potential inhibitors in several different studies, in particular flavanols quercetin and luteolin, which is not surprising as the most reported studies utilized similar target proteins and same software. Furthermore, the flavonoid core is present in many of putative inhibitors, suggesting that there is a shape complementarity between the active sites on the protein and flavonoid scaffold. The significance of these favourable interactions will be strengthened especially once the binding is validated and the dynamic behaviour of flavonoid-protein complexes is understood by using molecular dynamics.
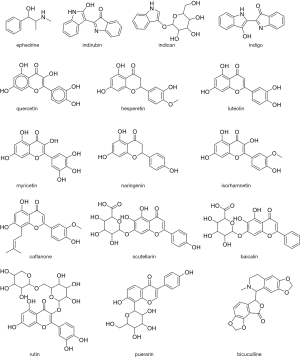
As the flavonoids are known to have poor bioavailability (64), these are not necessarily good drug candidates, but may provide a basis for design of analogues with acceptable ADME properties. As the more knowledge is acquired on the action of components of TCM and their formulations, the design of new molecules and formulations may become a “big data” that will allow use of machine learning approaches (65). Especially, emerging evidence about the use of TCM to treat COVID 19 (66,67) must be taken into account in future in silico studies. Moreover, the good practice of sharing the results of the studies should be further extended by supporting open drug discovery initiative as reported previously (68,69).
Finally, the validation of in silico results can only be achieved by testing most promising candidates in in vitro and by effective testing using animal models that are currently being developed (70).
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/lcm-20-42
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/lcm-20-42). MZ serves as an unpaid editorial board member of Longhua Chinese Medicine from Jul 2020 to Jun 2022. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- CDC. COVID-19 Response Team. Severe outcomes among patients with coronavirus disease 2019 (COVID-19)-United States, February 12-March 16, 2020. MMWR Morb Mortal Wkly Rep 2020;69:343-6. [Crossref] [PubMed]
- Spinelli A, Pellino G. COVID-19 pandemic: perspectives on an unfolding crisis. Br J Surg 2020;107:785-7. [Crossref] [PubMed]
- Nicola M, Alsafi Z, Sohrabi C, et al. The socio-economic implications of the coronavirus and COVID-19 pandemic: a review. Int J Surg 2020;78:185-93. [Crossref] [PubMed]
- Hsiang S, Allen D, Annan-Phan S, et al. The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature 2020;584:262-7. [Crossref] [PubMed]
- Pfefferbaum B, North CS. Mental health and the Covid-19 pandemic. N Engl J Med 2020;383:510-2. [Crossref] [PubMed]
- Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020;368:409-12. [Crossref] [PubMed]
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-3. [Crossref] [PubMed]
- Śledź P, Caflisch A. Protein structure-based drug design: from docking to molecular dynamics. Curr Opin Struct Biol 2018;48:93-102. [Crossref] [PubMed]
- Vanhaelen Q. Computational methods for drug repurposing. New York: Humana Press, 2019.
- Saxena A. Drug targets for COVID-19 therapeutics: ongoing global efforts. J Biosci 2020;45:87. [Crossref] [PubMed]
- Shyr ZA, Gorshkov K, Chen CZ, et al. Drug Discovery Strategies for SARS-CoV-2. J Pharmacol Exp Ther 2020;375:127-38. [Crossref] [PubMed]
- Panda PK, Arul MN, Patel P, et al. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci Adv 2020;6:eabb8097 [Crossref] [PubMed]
- Vijayakumar BG, Ramesh D, Joji A, et al. In silico pharmacokinetic and molecular docking studies of natural flavonoids and synthetic indole chalcones against essential proteins of SARS-CoV-2. Eur J Pharmacol 2020;886:173448 [Crossref] [PubMed]
- Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020;582:289-93. [Crossref] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61. [PubMed]
- Lin CW, Tsai FJ, Tsai CH, et al. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res 2005;68:36-42. [Crossref] [PubMed]
- Lin LT, Chung CY, Hsu WC, et al. Saikosaponin b2 is a naturally occurring terpenoid that efficiently inhibits hepatitis C virus entry. J Hepatol 2015;62:541-8. [Crossref] [PubMed]
- Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci 2020;29:1596-605. [Crossref] [PubMed]
- Sinha SK, Shakya A, Prasad SK, et al. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J Biomol Struct Dyn 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Gao LQ, Xu J, Chen SD. In silico screening of potential Chinese herbal medicine against COVID-19 by targeting SARS-CoV-2 3CLpro and angiotensin converting enzyme II using molecular docking. Chin J Integr Med 2020;26:527-32. [Crossref] [PubMed]
- Towler P, Staker B, Prasad SG, et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem 2004;279:17996-8007. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Jo S, Kim S, Shin DH, et al. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem 2020;35:145-51. [Crossref] [PubMed]
- Ngwa W, Kumar R, Thompson D, et al. Potential of flavonoid-inspired phytomedicines against COVID-19. Molecules 2020;25:2707. [Crossref] [PubMed]
- Fantini J, Di Scala C, Chahinian H, et al. Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents 2020;55:105960 [Crossref] [PubMed]
- Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res 2019;47:D948-54. [Crossref] [PubMed]
- Yao ZJ, Dong J, Che YJ, et al. TargetNet: a web service for predicting potential drug-target interaction profiling via multi-target SAR models. J Comput Aided Mol Des 2016;30:413-24. [Crossref] [PubMed]
- Wang X, Shen Y, Wang S, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 2017;45:W356-W360 [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Ge C, He Y. In Silico Prediction of molecular targets of astragaloside IV for alleviation of COVID-19 hyperinflammation by systems network pharmacology and bioinformatic gene expression analysis. Front Pharmacol 2020;11:556984 [Crossref] [PubMed]
- Wang SX, Wang Y, Lu YB, et al. Diagnosis and treatment of novel coronavirus pneumonia based on the theory of traditional Chinese medicine. J Integr Med 2020;18:275-83. [Crossref] [PubMed]
- Nicholls A, Grant JA. Molecular shape and electrostatics in the encoding of relevant chemical information. J Comput Aided Mol Des 2005;19:661-86. [Crossref] [PubMed]
- Chen H, Du Q. Potential natural compounds for preventing SARS-CoV-2 (2019-nCoV) infection. Preprints 2020. doi:
10.20944/preprints202001.0358.v3 . - Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol 2002;9:646-52. [Crossref] [PubMed]
- Huynh T, Wang H, Cornell W, et al. In Silico Exploration of Repurposing and Optimizing Traditional Chinese Medicine Rutin for Possibly Inhibiting SARS-CoV-2's Main Protease. ChemRxiv 2020. doi:
10.26434/chemrxiv.12281078.v1 . - Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci Rep 2016;6:21146. [Crossref] [PubMed]
- Kong R, Yang G, Xue R, et al. COVID-19 Docking Server: a meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Chen J, Wang YK, Gao Y, et al. Protection against COVID-19 injury by qingfei paidu decoction via anti-viral, anti-inflammatory activity and metabolic programming. Biomed Pharmacother 2020;129:110281 [Crossref] [PubMed]
- Yang R, Liu H, Bai C, et al. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): in silico and experimental study. Pharmacol Res 2020;157:104820 [Crossref] [PubMed]
- Xu HY, Zhang YQ, Liu ZM, et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res 2019;47:D976-82. [Crossref] [PubMed]
- Wu Y, Zhang F, Yang K, et al. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res 2019;47:D1110-7. [Crossref] [PubMed]
- Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res 2014;42:W32-8. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Ferrari A, Russo M. Introducing Microsoft Power BI. Redmond: Microsoft Press, 2016.
- Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45:D833-9. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Kanehisa M, Sato Y, Furumichi M, et al. New approach for understanding genome variations in KEGG. Nucleic Acids Res 2019;47:D590-5. [Crossref] [PubMed]
- Schriml LM, Mitraka E, Munro J, et al. Human Disease Ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res 2019;47:D955-62. [Crossref] [PubMed]
- Zhang DH, Zhang X, Peng B, et al. Network pharmacology suggests biochemical rationale for treating COVID-19 symptoms with a Traditional Chinese Medicine. Commun Biol 2020;3:466. [Crossref] [PubMed]
- Hong Z, Duan X, Wu S, et al. Network pharmacology integrated molecular docking reveals the anti-COVID-19 mechanism of Qing-Fei-Da-Yuan granules. Nat Prod Commun 2020;15:1934578X20934219.
- Han L, Wei XX, Zheng YJ, et al. Potential mechanism prediction of Cold-Damp Plague Formula against COVID-19 via network pharmacology analysis and molecular docking. Chin Med 2020;15:78. [Crossref] [PubMed]
- Yao YX, He ZX, Liu XF, et al. Potential material basis of Kangbingdu Granules for treatment of coronavirus disease 2019 (COVID-19) based on network pharmacology and molecular docking technology. Chin Trad Herbal Drugs 2020. doi:
10.7501/j.issn.0253-2670.2020.06.003 . - Zong Y, Ding ML, Jia KK, et al. Exploring active compounds of Da-Yuan-Yin in treatment of COVID-19 based on network pharmacology and molecular docking method. Chin Trad Herbal Drugs 2020. doi:
10.7501/j.issn.0253-2670.2020.04.002 . - Tao Q, Du J, Li X, et al. Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Huashi Baidu formula in the treatment of COVID-19. Drug Dev Ind Pharm 2020;46:1345-53. [Crossref] [PubMed]
- Zhao Y, Hu J, Song J, et al. Exploration on Shufeng Jiedu Capsule for Treatment of COVID-19 Based on Network Pharmacology and Molecular Docking. Chin Med 2020;11:9. [Crossref]
- Kong Y, Wu HW, Chen Y, et al. Mechanism of Tanreqing Injection on treatment of coronavirus disease 2019 based on network pharmacology and molecular docking. Chin Trad Herbal Drugs 2020:1785-94.
- Ruan X, Du P, Zhao K, et al. Mechanism of Dayuanyin in the treatment of coronavirus disease 2019 based on network pharmacology and molecular docking. Chin Med 2020;15:62. [Crossref] [PubMed]
- Selvaraj C, Dinesh DC, Panwar U, et al. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J Biomol Struct Dyn 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Ganesan A, Coote ML, Barakat K. Molecular dynamics-driven drug discovery: leaping forward with confidence. Drug Discov Today 2017;22:249-69. [Crossref] [PubMed]
- Jiang S, Cui Q, Ni B, et al. Databases for facilitating mechanistic investigations of traditional Chinese medicines against COVID-19. Pharmacol Res 2020;159:104989 [Crossref] [PubMed]
- Huang YX, Wang WX, Zhang S, et al. The database-based strategy may overstate the potential effects of traditional Chinese medicine against COVID-19. Pharmacol Res 2020;159:105046 [Crossref] [PubMed]
- Thilakarathna SH, Rupasinghe H. Flavonoid bioavailability and attempts for bioavailability enhancement. Nutrients 2013;5:3367-87. [Crossref] [PubMed]
- Zloh M, Kirton SB. The benefits of in silico modeling to identify possible small-molecule drugs and their off-target interactions. Future Med Chem 2018;10:423-32. [Crossref] [PubMed]
- Luo H, Gao Y, Zou J, et al. Reflections on treatment of COVID-19 with traditional Chinese medicine. Chin Med 2020;15:94. [Crossref] [PubMed]
- Fan L, Jiang S, Yang X, et al. COVID-19 Drug Treatment in China. Curr Pharmacol Rep 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Ekins S, Mottin M, Ramos PRPS, et al. Déjà vu: Stimulating open drug discovery for SARS-CoV-2. Drug Discov Today 2020;25:928-41. [Crossref] [PubMed]
- Chodera J, Lee AA, London N, et al. Crowdsourcing drug discovery for pandemics. Nat Chem 2020;12:581. [Crossref] [PubMed]
- Muñoz-Fontela C, Dowling WE, Funnell SGP, et al. Animal models for COVID-19. Nature 2020;586:509-15. [Crossref] [PubMed]
Cite this article as: Zloh M, Ilić M, Jojić N, Gigov S, Jovanović Lješković N. Narrative review of the progress in discovery of anti COVID-19 agents from traditional Chinese medicine using in silico approaches. Longhua Chin Med 2020;3:19.

