
Adaptogens in chemobrain (part IV): adaptogenic plants prevent the chemotherapeutics-induced imbalance of redox homeostasis by modulation of expression of genes encoding Nrf2-mediated signaling proteins and antioxidant, metabolizing, detoxifying enzymes in neuroglia cells
Introduction
Several plants widely used in traditional Chinese medicine (TCM), Ayurveda and other traditional medicinal systems, such as Panax ginseng Meyer root, Eleutherococcus senticosus (Rupr. & Maxim.) Maxim. root (ES), Rhodiola rosea L. rhizome and root (RR), Andrographis paniculata L. Nees herb (AP), and Schisandra chinensis (Turcz.) Bail. (berry) (SC), are known for their pleiotropic effects, which are considered to be associated with the adaptogenic activity; that is, an ability to increase adaptability (presumably “xiehuo” in TCM), resilience, and survival of organisms in stress- and aging-related disorders (1-5). This could be due to increasing the “state of non-specific resistance” (6-8), which is negatively associated with the state of increased susceptibility to stressors—“shanghuo” or “re-qi” in TCM—resulting in disease progression due to threatened homeostasis (yin-yang balance) (9,10).
The neuroprotective, hepatoprotective, cardioprotective, antioxidant, immunomodulatory, antiviral, stress-protective, antifatigue, hypoglycemic, antidepressant, chemopreventive, and antitoxic effects of various adaptogenic preparations have been shown in many isolated cells and in experimental animal models (11-35).
The cytoprotective effect of andrographolide (AND; an active compound of AP), AP, ES, AP-ES combination, and ES-RR-SC combination on the chemotherapy-induced deregulation of gene expression in neuroglia cell culture has been recently demonstrated (36-38). These studies suggest that adaptogens might be useful for preventing and mitigating the toxic effect of chemotherapy in cancer (e.g., in “chemobrain”) (39), but also for reducing oxidative stress-induced cellular damage and detoxification in many inflammatory conditions, including low-grade chronic inflammation (“inflammaging”) in aging (40). Oxidative stress is increasing in aging-related disorders, including atherosclerosis, angiogenesis, and neurodegeneration (40,41). Oxidative stress-induced redox signaling results in cellular response and the activation of defense mechanisms, including the induction of antioxidant and detoxifying enzymes and molecular chaperones (42). Nuclear factor erythroid 2-related factor 2 (Nrf2) is a principal regulator of redox homeostasis, triggering the expression of antioxidant and detoxifying genes, including superoxide dismutase (SOD), glutathione S-transferase (GST), NAD(P)H quinone oxidoreductase-1 (NQO1), and heme oxygenase 1 (HO-1). Therefore, the activation of Nrf2 translocation or the upregulation of gene expression resulting in the activation of the Nrf2 signaling pathway is the key mechanism of cellular defense response associated with the antioxidant and detoxifying effects of medicinal plants (43-46), and particularly of adaptogenic plants, such as AP, RR, SC, and ES (47-71). However, their effect on the expression of genes encoding antioxidant enzymes, phase II and III metabolizing enzymes, and transports, as well on upstream transcription factors, has not been investigated. Therefore, the aim of the present study was to assess the effects of AND, AP, ES, AP-ES combination, and ES-RR-SC combination on chemotherapy [fixed combination 5-fluorouracil, epirubicin, and cyclophosphamide (FEC)]-induced gene expression related to Nrf2 signaling pathways in neuroglia T98G cell culture.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/lcm-20-24).
Methods
All materials and methods used in the present study have been described in detail in our previously published studies (36-38). The present study is a part of the same study; however, the results have not been published until now. Therefore, only short description of herbal extracts, mRNA microarray hybridization and ingenuity pathway analysis (IPA) is provided below.
Herbal extracts
Pharmaceutical-grade extracts were manufactured in accordance with the ICHQ7A and European Medical Agency guidelines for good agricultural and collection practice and good manufacturing practice of active pharmaceutical ingredients. Working samples used in the experiments were prepared by diluting dimethylsulfoxide (DMSO) solutions of the extracts with appropriate volumes of phosphate-buffered saline to obtain the same final concentrations of active markers in the incubation media.
The concentrations of the extracts and their active constituents were well-matched in all test samples; that is, the final concentration of AND was the same (2 µm, 700 µg/mL) in all test samples containing AND, and corresponded to the concentration of AND in human blood after the oral administration of a therapeutic dose (60 mg) of herbal (Table 1) (71). Similarly, eleuthero side E concentrations were calculated based on the results of HPLC analyses of its content in genuine extracts and AP-ES, ES-RR-SC combinations. The concentrations of genuine extracts were calculated using AE specifications to ensure that they corresponded to therapeutically effective doses (35).
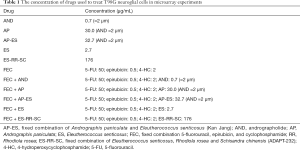
Full table
mRNA microarray hybridization
T98G cells were seeded and attached for 24 hours prior to drug treatment. Cells were treated for 24 hours at various combinations and concentrations of drugs or DMSO as the solvent control (0.5%). Total RNA was then isolated using the InviTrap Spin Universal RNA mini kit (250 preps; Stratec Molecular, Germany). RNA concentrations were determined using the NanoDrop spectrophotometer (NanoDrop Technologies, USA). The quality of total RNA was confirmed by gel analysis using the total RNA Nanochip assay on an Agilent 2100 bioanalyzer (Agilent Technologies GmbH, USA). Only samples with RNA index values >8.5 were selected for expression profiling. The experiment was performed in duplicate for treated samples and for control samples by the Genomics and Proteomics Core Facility at the German Cancer Research Center in Heidelberg, Germany. Biotin-labeled cRNA samples for hybridization on Illumina Human HT-12 v4 BeadChip arrays were prepared according to Illumina’s recommended sample labeling procedure based on the modified Eberwine protocol. In brief, 250–500 ng total RNA was used for cDNA synthesis, followed by an amplification/labeling step (in vitro transcription) to synthesize biotin-labeled cRNA according to the MessageAmp II aRNA amplification kit (Ambion, USA). Biotin-16-UTP was purchased from Roche Applied Science. The cRNA was column purified according to the Total Prep RNA amplification kit and eluted in 60–80 µL water. The cRNA quality was controlled using the RNA nanochip assay on an Agilent 2100 bioanalyzer and was spectrophotometrically quantified (NanoDrop). Subsequent hybridization was performed according to the manufacturer’s instruction. Microarray scanning was done using a Beadstation array scanner with the setting adjusted to a scaling factor of 1 and a photomultiplier tube setting of 430. Data extraction was performed for all beads individually, and outliers were removed when the median absolute deviation exceeded 2.5. The mean average signals and standard deviations (SDs) were then calculated for each probe. Data analysis was done by normalization of the signals using the quantile normalization algorithm without background subtraction, and differentially regulated genes were defined by calculating the SD differences of a given probe in a one-by-one comparison of samples or groups. The data were further processed using Chipster software (The Finnish IT Center for Science CSC).
IPA
Microarray data were analyzed by IPA (Ingenuity Systems, USA). IPA software relies on the Ingenuity Knowledge Base, a frequently updated database containing biologic and chemical interactions and functional annotations gathered from the literature. In order to obtain information about cellular functions, networks, and affected pathways, IPA offers the Core Analysis tool, which was used for all datasets.
IPA performs different calculations on transcriptomic datasets, including prediction algorithms, and produces results of analyses in a variety of ways, including (I) canonical pathways, which displays the molecules of interest within well-established signaling or metabolic pathways; and (II) upstream analysis, which predicts the upstream regulators (any molecule that can influence the transcription or expression of another molecule) that might be activated or inhibited to explain the expression changes in test datasets.
The interpretation of gene expression data was facilitated by consideration of prior biologic knowledge. IPA software relies on the Ingenuity Knowledge Base, a large gathering of observations with more than 5 million findings manually curated from the biomedical literature or integrated from third-party databases. The network contains 40,000 nodes that represent mammalian genes, molecules, and biologic functions. Nodes are linked by 1,480,000 edges representing experimentally observed cause-effect relationships that relate to gene expression, transcription, activation, molecular metabolism, and binding. Network edges are also associated with a direction (either activating or inhibiting) of the causal effect (72).
To obtain information about the impact of test samples on cellular signaling pathways and networks, for biologic functions and diseases downstream of the genes, whose expression has been altered in a dataset, we used the IPA Core Analysis tool for all tested datasets. Analysis of transcriptomics enabled us to predict regulators that are activated or inhibited based on the distinct up- and downregulation patterns of the expressed genes, and to determine which causal relationships previously reported in the literature are likely to be relevant for the biologic mechanisms underlying the data.
Statistical analysis
Two statistical methods of analysis of gene expression data were used in IPA: (I) gene-set-enrichment method, where differentially expressed genes are intersected with sets of genes that are associated with a particular biological function or pathway providing an ‘enrichment’ score (Fisher’s exact test P value) that measures overlap of observed and predicted regulated gene sets (73,74); (II) The method that based on previously observed cause-effect relationships related to the direction of effects reported in the literature (75,76) providing so called Z-scores assessing the match of observed and predicted up/down regulation patterns (72). The predicted [Z-score >2; or –log (FET P value) >1.3] effects are based on changes of gene expression in the experimental samples relative to the control.
Results
Microarray-based, transcriptome-wide mRNA expression analyses were performed to identify possible targets of the FEC, herbal extracts, and their fixed combination in T98G cells.
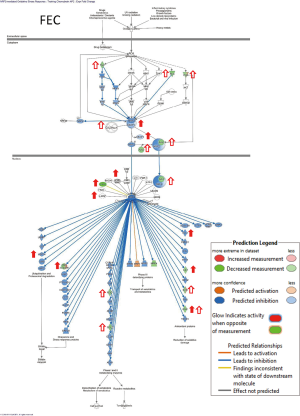
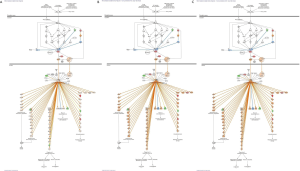
FEC significantly (>two-fold) deregulated 23 genes of Nrf2 signaling pathways (Figure 1). Co-incubation with adaptogens induces the upregulation of HMOX1 expression and genes encoding cytoplasmatic transcription factor Kelch-like ECH-associated protein (KEAP1), a key protein involved in the activation of KEAP1-Nrf2-mediated signaling and nuclear transcription factors MAF bZIP transcription factor F and MAF bZIP transcription factor G (Table 2). Moreover, adaptogens prevent FEC-induced downregulation of genes PIK3R2 and RALA encoding Nrf2 upstream signaling proteins PI3K and/Ras, as well as genes NQO1, GSR, and GCLC, encoding the expression of cytoplasmatic antioxidant enzymes glutathione-disulfide reductase, NAD(P)H quinone dehydrogenase 1, and glutamate-cysteine ligase catalytic subunit (Table 2). The effects on other gene expressions of Nrf2-mediated signaling are shown in Figure 1 and Table 2.

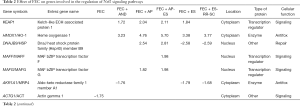
Full table
Discussion
Nrf2 is a principal regulator of redox homeostasis, normally retained in the cytoplasm by KEAP1 (42,77-79). Upon exposure of cells to oxidative stress, Nrf2 is phosphorylated in response to protein kinase C, phosphatidylinositol 3-kinase, and the mitogen-activated protein kinase pathways. After phosphorylation, this complex dissociates, and Nrf2 translocates to the nucleus where it binds with the antioxidant response element (ARE) and triggers the expression of antioxidant and detoxifying genes, including SOD, GST, NQO1, and HMOX1 (42,77). Therefore, the activation of Nrf2 translocation or the upregulation of gene expression resulting in the activation of the Nrf2 mediated signaling pathway is the key mechanism of cellular defense response associated with the antioxidant effects of medicinal plants (43-45), and particularly of adaptogenic plants, which are useful in stress- and aging-related diseases (40,41,78).
Figures 1,2 show that FEC inhibits the Nrf2 signaling pathway via the deregulation of expression of 24 genes, whereas adaptogens prevent or mitigate FEC-induced deregulation of a number of genes involved in the predicted activation of Nrf2-mediated signaling and expression of antioxidant and detoxifying genes, including SOD, GST, NQO1, and HMOX1 (Figures 3,4).
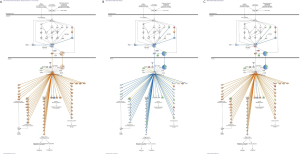
Conclusions
The results of the present study suggest that the beneficial effects of adaptogens on impaired neuronal and cognitive functions are due to mitigating oxidative stress-induced cellular damage by multitarget regulation of redox homeostasis via the regulation of gene expression, activating Nrf2 signaling pathway proteins and modulating antioxidant, metabolizing, and detoxifying enzymes.
Acknowledgments
The authors acknowledge the support of the Swedish Herbal Institute for supplying the investigational agents used in the present study.
Funding: The present study was partially funded by the Swedish Herbal Institute.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/lcm-20-24
Data Sharing Statement: Available at http://dx.doi.org/10.21037/lcm-20-24
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/lcm-20-24). AP serves as an unpaid editorial board member of Longhua Chinese Medicine from May 2020 to Apr 2022. AP is the head of research and development company Phytomed AB and has an independent contractor agreement with EuroPharma USA. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). No ethic committee’s approval is required in Germany for
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Panossian A. Understanding adaptogenic activity: specificity of the pharmacological action of adaptogens and other phytochemicals. Ann NY Acad Sci 2017;1401:49-64. [Crossref] [PubMed]
- Panossian A, Seo EJ, Efferth T. Novel molecular mechanisms for the adaptogenic effects of herbal extracts on isolated brain cells using systems biology. Phytomedicine 2018;50:257-84. [Crossref] [PubMed]
- Panossian AG. Adaptogens: tonic herbs for fatigue and stress. Alternative & Complementary Therapies 2003;9:327-32. [Crossref]
- Wagner H, Nörr H, Winterhoff H. Plant adaptogens. Phytomedicine 1994;1:63-76. [Crossref] [PubMed]
- Panossian A, Wagner H. Adaptogens. A review of their history, biological activity, and clinical benefits. HerbalGram 2011;90:52-63.
- Lazarev NV. General and specific in action of pharmacological agents. Farmacol Toxicol 1958;21:81-6.
- Lazarev NV, Liublina EI, Rozin MA. States of non-specific increased resistance. Patol Fiziol Eksp Ter 1959;3:16-21. [PubMed]
- Brekhman II, Dardymov IV. New substances of plant origin which increase nonspecific resistance. Annu Rev Pharmacol 1969;9:419-30. [Crossref] [PubMed]
- He RR, Hiroshi K. Shanghuo syndrome in traditional Chinese medicine. World Sci Technol 2008;10:37-41. [Crossref]
- Pan MH, Zhu SR, Duan WJ, et al. "Shanghuo" increases disease susceptibility: Modern significance of an old TCM theory. J Ethnopharmacol 2020;250:112491 [Crossref] [PubMed]
- EMA/HMPC/232100/2011. Assessment report on Rhodiola rosea L., rhizoma et radix. Based on Article 16d(1), Article 16f and Article 16h of Directive 2001/83/EC as amended traditional use). Final. 27 March 2012.
- EMA/HMPC/321232/2012. Assessment report on Panax ginseng C.A. Meyer, radix. Based on Article 16d (1), Article 16f and Article 16h of Directive 2001/83/EC as amended (traditional use). Final. 25 March 2014.
- EMA/HMPC/680615/2013. Assessment report on Eleutherococcus senticosus (Rupr. et Maxim.) Maxim., radix. Based on Article 16d(1), Article 16f and Article 16h of Directive 2001/83/EC as amended (traditional use). Final. 25 March 2014.
- EMA/HMPC/320433/2012. Assessment report on Andrographis paniculata Nees, folium. Based on Article 10a of Directive 2001/83/EC as amended (well-established use). Based on Article 16d (1), Article 16f and Article 16h of Directive 2001/83/EC as amended (traditional use). Final. 27 August 2014.
- Farnsworth NR, Waller D, Strelkova LM. Use of Eleutherococcus senticosus in United States: problems, prospects and literature update. Vladivostok: Proceedings of the Second International Symposium on Eleutherococcus, 1986:47-51.
- Panossian AG. Adaptogens in mental and behavioral disorders. Psychiatr Clin North Am 2013;36:49-64. [Crossref] [PubMed]
- Panossian A, Wikman G. Effects of adaptogens on the central nervous system and the molecular mechanisms associated with their stress-protective activity. Pharmaceuticals (Basel) 2010;3:188-224. [Crossref] [PubMed]
- Panossian A, Wikman G. Pharmacology of Schisandra chinensis Bail.: an overview of Russian research and uses in medicine. J Ethnopharmacol 2008;118:183-212. [Crossref] [PubMed]
- Thakur AK, Chatterjee SS, Kumar V. Adaptogenic potential of andrographolide: an active principle of the king of bitters (Andrographis paniculata). J Tradit Complement Med 2014;5:42-50. [Crossref] [PubMed]
- Thakur AK, Soni UK, Rai G, et al. Protective effects of Andrographis paniculata extract and pure andrographolide against chronic stress-triggered pathologies in rats. Cell Mol Neurobiol 2014;34:1111-21. [Crossref] [PubMed]
- Bertoglio JC, Baumgartner M, Palma R, et al. Andrographis paniculata decreases fatigue in patients with relapsing-remitting multiple sclerosis: a 12-month double-blind placebo-controlled pilot study. BMC Neurol 2016;16:77. [Crossref] [PubMed]
- Brekhman II. On antitoxic action of Eleutherococcus. Moscow: Meditsina, 1982.
- Lin CC, Huang PC. Antioxidant and hepatoprotective effects of Acanthopanax senticosus. Phytother Res 2000;14:489-94. [Crossref] [PubMed]
- Monokhov BV. Influence of the liquid extract from the roots of Eleutherococcus senticosus on the toxicity and antitumor activity of cyclophosphan. Vopr Onkol 1965;11:60-3. [PubMed]
- Sakharova TA, Revazova YA, Barenboim GM. The effect of Eleutherococcus extract on the induction of recessive lethal mutations by cyclophosphane and N-nitrosomorpholine in Drosophila. Khimiko Farmatseevticheskii Zhurnal 1985;19:539-40.
- Smalinskiene A, Lesauskaite V, Zitkevicius V, et al. Estimation of the combined effect of Eleutherococcus senticosus extract and cadmium on liver cells. Ann N Y Acad Sci 2009;1171:314-20. [Crossref] [PubMed]
- Park SH, Lee SG, Kang SK, et al. Acanthopanax senticosus reverses fatty liver disease and hyperglycemia in ob/ob mice. Arch Pharm Res 2006;29:768-76. [Crossref] [PubMed]
- Maslov LN, Guzarova NV. Cardioprotective and antiarrhythmic properties of preparations from Leuzea carthamoides, Aralia mandshurica, and Eleutherococcus senticosus. Eksp Klin Farmakol 2007;70:48-54. [PubMed]
- Maslov LN, Lishmanov YB, Arbuzov AG, et al. Antiarrhythmic activity of phytoadaptogens in short-term ischemia-reperfusion of the heart and postinfarction cardiosclerosis. Bull Exp Biol Med 2009;147:331-4. [Crossref] [PubMed]
- Sheeja K, Kuttan G. Ameliorating effects of Andrographis paniculata extract against cyclophosphamide-induced toxicity in mice. Asian Pac J Cancer Prev 2006;7:609-14. [PubMed]
- Singh P, Srivastava MM, Khemani LD. Renoprotective effects of Andrographis paniculata (Burm. f.) Nees in rats. Ups J Med Sci 2009;114:136-9. [Crossref] [PubMed]
- Gupta S, Mishra KP, Singh SB, et al. Inhibitory effects of andrographolide on activated macrophages and adjuvant-induced arthritis. Inflammopharmacology 2018;26:447-56. [Crossref] [PubMed]
- Islam MT. Andrographolide, a new hope in the prevention and treatment of metabolic syndrome. Front Pharmacol 2017;8:571. [Crossref] [PubMed]
- Jayakumar T, Hsieh CY, Lee JJ, et al. Experimental and clinical pharmacology of Andrographis paniculata and its major bioactive phytoconstituent andrographolide. Evid Based Complement Alternat Med 2013;2013:846740 [Crossref] [PubMed]
- Panossian A, Wikman G. Efficacy of Andrographis paniculata in upper respiratory tract (URT) infectious diseases and the mechanism of action. In: Wagner H, Ulrich-Merzenich G. editors. Evidence and rational based research on Chinese Drugs. Vienna: Springer, 2013:137-80.
- Seo EJ, Klauck SM, Efferth T, et al. Adaptogens in chemobrain (part I): plant extracts attenuate cancer chemotherapy-induced cognitive impairment - Transcriptome-wide microarray profiles of neuroglia cells. Phytomedicine 2019;55:80-91. [Crossref] [PubMed]
- Seo EJ, Klauck SM, Efferth T, et al. Adaptogens in chemobrain (Part II): effect of plant extracts on chemotherapy-induced cytotoxicity in neuroglia cells. Phytomedicine 2019;58:152743 [Crossref] [PubMed]
- Seo EJ, Klauck SM, Efferth T, et al. Adaptogens in chemobrain (part III): antitoxic effects of plant extracts towards cancer chemotherapy-induced toxicity - transcriptome-wide microarray analysis of neuroglia cells. Phytomedicine 2019;56:246-60. [Crossref] [PubMed]
- O'Farrell E, MacKenzie J, Collins B. Clearing the air: a review of our current understanding of "Chemo Fog". Curr Oncol Rep 2013;15:260-9. [Crossref] [PubMed]
- Franceschi C, Garagnani P, Parini P, et al. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 2018;14:576-90. [Crossref] [PubMed]
- Li M, Fukagawa NK. Age-related changes in redox signaling and VSMC function. Antioxid Redox Signal 2010;12:641-55. [Crossref] [PubMed]
- Hybertson BM, Gao B, Bose SK, et al. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol Aspects Med 2011;32:234-46. [Crossref] [PubMed]
- Reuland DJ, McCord JM, Hamilton KL. The role of Nrf2 in the attenuation of cardiovascular disease. Exerc Sport Sci Rev 2013;41:162-8. [Crossref] [PubMed]
- Ajit D, Simonyi A, Li R, et al. Phytochemicals and botanical extracts regulate NF-κB and Nrf2/ARE reporter activities in DI TNC1 astrocytes. Neurochem Int 2016;97:49-56. [Crossref] [PubMed]
- Bjørklund G, Dadar M, Martins N, et al. Brief challenges on medicinal plants: an eye-opening look at ageing-related disorders. Basic Clin Pharmacol Toxicol 2018;122:539-58. [Crossref] [PubMed]
- Adeoye BO, Asenuga ER, Oyagbemi AA, et al. The protective effect of the ethanol leaf extract of Andrographis paniculata on cisplatin-induced acute kidney injury in rats through nrf2/KIM-1 signalling pathway. Drug Res (Stuttg) 2018;68:23-32. [Crossref] [PubMed]
- Pan CW, Yang SX, Pan ZZ, et al. Andrographolide ameliorates d-galactosamine/lipopolysaccharide-induced acute liver injury by activating Nrf2 signaling pathway. Oncotarget 2017;8:41202-10. [Crossref] [PubMed]
- Seo JY, Pyo E, An JP, et al. Andrographolide activates Keap1/Nrf2/ARE/HO-1 pathway in HT22 cells and suppresses microglial activation by Aβ(42) through Nrf2-related inflammatory response. Mediators Inflamm 2017;2017:5906189 [Crossref] [PubMed]
- Wong SY, Tan MG, Wong PT, et al. Andrographolide induces Nrf2 and heme oxygenase 1 in astrocytes by activating p38 MAPK and ERK. J Neuroinflammation 2016;13:251. [Crossref] [PubMed]
- Lin HC, Su SL, Lu CY, et al. Andrographolide inhibits hypoxia-induced HIF-1α-driven endothelin 1 secretion by activating Nrf2/HO-1 and promoting the expression of prolyl hydroxylases 2/3 in human endothelial cells. Environ Toxicol 2017;32:918-30. [Crossref] [PubMed]
- Lu CY, Yang YC, Li CC, et al. Andrographolide inhibits TNFα-induced ICAM-1 expression via suppression of NADPH oxidase activation and induction of HO-1 and GCLM expression through the PI3K/Akt/Nrf2 and PI3K/Akt/AP-1 pathways in human endothelial cells. Biochem Pharmacol 2014;91:40-50. [Crossref] [PubMed]
- Lee JC, Tseng CK, Young KC, et al. Andrographolide exerts anti-hepatitis C virus activity by up-regulating haeme oxygenase-1 via the p38 MAPK/Nrf2 pathway in human hepatoma cells. Br J Pharmacol 2014;171:237-52. [Crossref] [PubMed]
- Yuan XY, Pang XW, Zhang GQ, et al. Salidroside's protection against UVB-mediated oxidative damage and apoptosis is associated with the upregulation of Nrf2 expression. Photomed Laser Surg 2017;35:49-56. [Crossref] [PubMed]
- Zhu Y, Zhang YJ, Liu WW, et al. Salidroside suppresses HUVECs cell injury induced by oxidative stress through activating the Nrf2 signaling pathway. Molecules 2016;21:1033. [Crossref] [PubMed]
- Han J, Xiao Q, Lin YH, et al. Neuroprotective effects of salidroside on focal cerebral ischemia/reperfusion injury involve the nuclear erythroid 2-related factor 2 pathway. Neural Regen Res 2015;10:1989-96. [Crossref] [PubMed]
- Tang H, Gao L, Mao J, et al. Salidroside protects against bleomycin-induced pulmonary fibrosis: activation of Nrf2-antioxidant signaling, and inhibition of NF-κB and TGF-β1/Smad-2/-3 pathways. Cell Stress Chaperones 2016;21:239-49. [Crossref] [PubMed]
- Kwon DH, Cha HJ, Choi EO, et al. Schisandrin A suppresses lipopolysaccharide-induced inflammation and oxidative stress in RAW 264.7 macrophages by suppressing the NF-κB, MAPKs and PI3K/Akt pathways and activating Nrf2/HO-1 signaling. Int J Mol Med 2018;41:264-74. [PubMed]
- Kim EJ, Jang M, Lee MJ, et al. Schisandra chinensis stem ameliorates 3-nitropropionic acid-induced striatal toxicity via activation of the nrf2 pathway and inhibition of the MAPKs and NF-κB pathways. Front Pharmacol 2017;8:673. [Crossref] [PubMed]
- Lai Q, Luo Z, Wu C, et al. Attenuation of cyclosporine A induced nephrotoxicity by schisandrin B through suppression of oxidative stress, apoptosis and autophagy. Int Immunopharmacol 2017;52:15-23. [Crossref] [PubMed]
- Chen Q, Zhang H, Cao Y, et al. Schisandrin B attenuates CCl(4)-induced liver fibrosis in rats by regulation of Nrf2-ARE and TGF-β/Smad signaling pathways. Drug Des Devel Ther 2017;11:2179-91. [Crossref] [PubMed]
- Lin Q, Qin X, Shi M, et al. Schisandrin B inhibits LPS-induced inflammatory response in human umbilical vein endothelial cells by activating Nrf2. Int Immunopharmacol 2017;49:142-7. [Crossref] [PubMed]
- Jiang YM, Wang Y, Tan HS, et al. Schisandrol B protects against acetaminophen-induced acute hepatotoxicity in mice via activation of the NRF2/ARE signaling pathway. Acta Pharmacol Sin 2016;37:382-9. [Crossref] [PubMed]
- He JL, Zhou ZW, Yin JJ, et al. Schisandra chinensis regulates drug metabolizing enzymes and drug transporters via activation of Nrf2-mediated signaling pathway. Drug Des Devel Ther 2014;9:127-46. [PubMed]
- Kang JS, Han MH, Kim GY, et al. Nrf2-mediated HO-1 induction contributes to antioxidant capacity of a Schisandrae Fructus ethanol extract in C2C12 myoblasts. Nutrients 2014;6:5667-78. [Crossref] [PubMed]
- Kang JS, Han MH, Kim GY, et al. Schisandrae semen essential oil attenuates oxidative stress-induced cell damage in C2C12 murine skeletal muscle cells through Nrf2-mediated upregulation of HO-1. Int J Mol Med 2015;35:453-9. [Crossref] [PubMed]
- Checker R, Patwardhan RS, Sharma D, et al. Schisandrin B exhibits anti-inflammatory activity through modulation of the redox-sensitive transcription factors Nrf2 and NF-κB. Free Radic Biol Med 2012;53:1421-30. [Crossref] [PubMed]
- Lee SB, Kim CY, Lee HJ, et al. Induction of the phase II detoxification enzyme NQO1 in hepatocarcinoma cells by lignans from the fruit of Schisandra chinensis through nuclear accumulation of Nrf2. Planta Med 2009;75:1314-8. [Crossref] [PubMed]
- Zhang A, Liu Z, Sheng L, et al. Protective effects of syringin against lipopolysaccharide-induced acute lung injury in mice. J Surg Res 2017;209:252-7. [Crossref] [PubMed]
- Jin ML, Park SY, Kim YH, et al. Acanthopanax senticosus exerts neuroprotective effects through HO-1 signaling in hippocampal and microglial cells. Environ Toxicol Pharmacol 2013;35:335-46. [Crossref] [PubMed]
- Wang X, Hai CX, Liang X, et al. The protective effects of Acanthopanax senticosus Harms aqueous extracts against oxidative stress: role of Nrf2 and antioxidant enzymes. J Ethnopharmacol 2010;127:424-32. [Crossref] [PubMed]
- Panossian A, Hovhannisyan A, Mamikonyan G, et al. Pharmacokinetic and oral bioavailability of andrographolide from Andrographis paniculata fixed combination Kan Jang in rats and human. Phytomedicine 2000;7:351-64. [Crossref] [PubMed]
- Krämer A, Green J, Pollard J Jr, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523-30. [Crossref] [PubMed]
- Abatangelo L, Maglietta R, Distaso A, et al. Comparative study of gene set enrichment methods. BMC Bioinformatics 2009;10:275. [Crossref] [PubMed]
- Ihnatova I, Popovici V, Budinska E. A critical comparison of topology-based pathway analysis methods. PLoS One 2018;13:e0191154 [Crossref] [PubMed]
- Chindelevitch L, Ziemek D, Enayetallah A, et al. Causal reasoning on biological networks: interpreting transcriptional changes. Bioinformatics 2012;28:1114-21. [Crossref] [PubMed]
- Fakhry CT, Choudhary P, Gutteridge A, et al. Interpreting transcriptional changes using causal graphs: new methods and their practical utility on public networks. BMC Bioinformatics 2016;17:318. [Crossref] [PubMed]
- Tebay LE, Robertson H, Durant ST, et al. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med 2015;88:108-46. [Crossref] [PubMed]
- Deshmukh P, Unni S, Krishnappa G, et al. The Keap1-Nrf2 pathway: promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys Rev 2017;9:41-56. [Crossref] [PubMed]
- Ahmed SM, Luo L, Namani A, et al. Nrf2 signaling pathway: pivotal roles in inflammation. Biochim Biophys Acta Mol Basis Dis 2017;1863:585-97. [Crossref] [PubMed]
(English Language Editors: R. Scott and J. Chapnick)
Cite this article as: Panossian A, Seo EJ, Klauck SM, Efferth T. Adaptogens in chemobrain (part IV): adaptogenic plants prevent the chemotherapeutics-induced imbalance of redox homeostasis by modulation of expression of genes encoding Nrf2-mediated signaling proteins and antioxidant, metabolizing, detoxifying enzymes in neuroglia cells. Longhua Chin Med 2020;3:4.

